Radial or angular distribution function (RDF, ADF)¶
RDF¶
RDF is commonly used to investigate structure of the system, e.g., solid vs liquid, or crystal vs amorphous.
To get the RDF,
python /path/to/nap/nappy/rdf.py [options] dump_0*
then, you get averaged RDF over atoms in out.rdf.
Given atom configuration files, dump_####, are read and average over
atoms in those files are taken.
Options are shown below,
Options:
-h, --help Show this help message and exit.
-d DR Width of the bin. [default: 0.1]
-r RMAX Cutoff radius of radial distribution. [default: 5.0]
--gsmear=SIGMA
Width of Gaussian smearing, zero means no smearing. [default: 0]
-o OUT Output file name. [default: out.rdf]
--specorder=SPECORDER
Order of species separated by comma, like, --specorder=W,H. [default: None]
--skip=NSKIP
Skip first NSKIP steps from the statistics. [default: 0]
--no-average
Not to take average over files.
--no-normalize
Not to normalize by the density.
--plot Plot figures. [default: False]
The RDF of each pair of species normalized with the density of the pair.
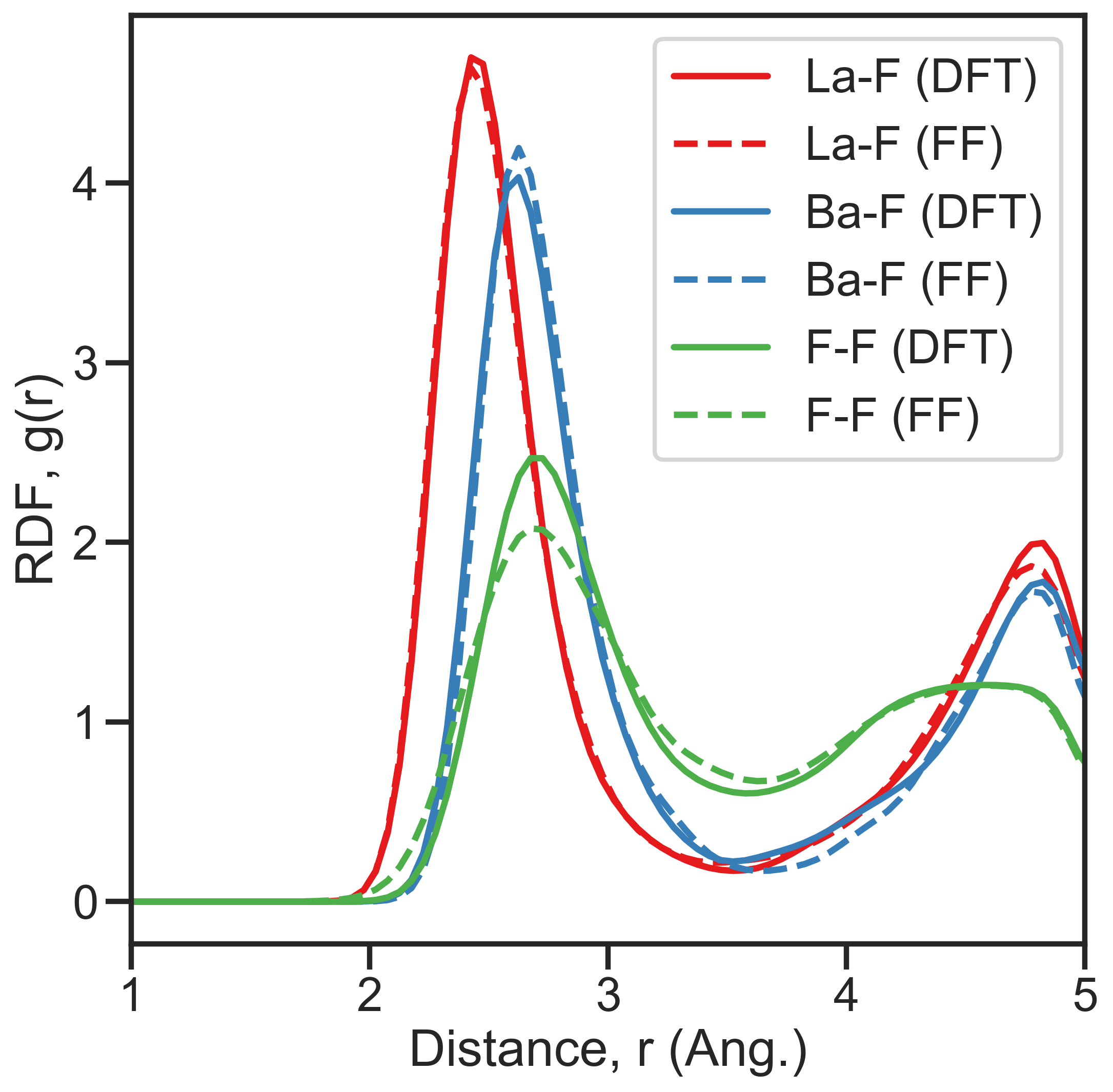
ADF¶
To get ADF, perform adf.py something like,
python /path/to/nap/nappy/adf.py --triplets=La-F-F,Ba-F-F dump_0*
The triplets consisting angles must be provided via the option
--triplets. Note that the 1st species in the triplet is the central
atom having bonds to the other two atoms, which maybe counter-intuitive.
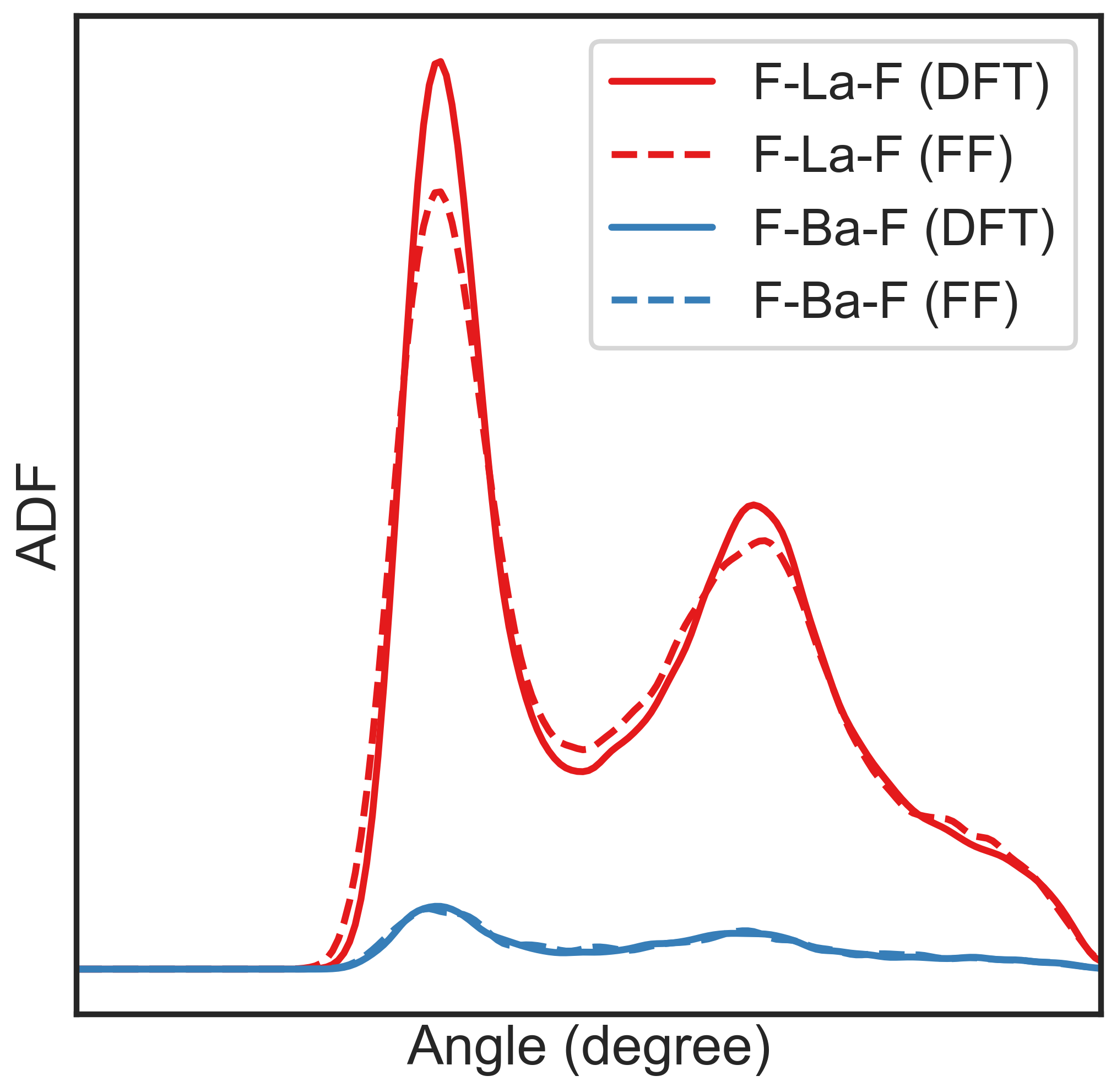
Here is some options of adf.py,
Options:
-h, --help Show this help message and exit.
-w DEG Width of the angular degree. [default: 1.0]
-r RCUT Cutoff radius of the bonding pair. [default: 3.0]
--gsmear=SIGMA
Width of Gaussian smearing, zero means no smearing. [default: 0]
--triplets=TRIPLETS
Triplets whose angles are to be computed. Three species should be specified connected by hyphen,
and separated by comma, e.g.) P-O-O,Li-O-O. [default: None]
-o OUT Output file name [default: out.adf]
--skip=NSKIP
Skip first NSKIP steps from the statistics. [default: 0]
--no-average
Not to take average over files.
--plot Plot figures. [default: False]